Monday February 26, 2024

Humans around the world rely heavily on freshwater biodiversity for healthy ecosystems, food, and income. However, this aquatic wealth is under threat – as evidenced by recent reports that as much as a quarter of all known freshwater fish are at significant risk of extinction. These alarming declines are particularly concerning in hyper-diverse tropical river systems like the Mekong, which is home to over 1,000 species of fish. Monitoring biodiversity in a basin as large and diverse as the Mekong represents a substantial challenge, but new technologies stand poised to aid in this Herculean task. Among these is environmental DNA (eDNA) sampling, which allows scientists to detect fish species by taking a water sample. However, much work remains to be done to establish best practices for eDNA in tropical rivers. To this end, FISHBIO and partners from Young Eco conducted a pilot study funded by the US Agency for International Development (USAID) Wonders of the Mekong project to evaluate eDNA’s ability to quantify fish diversity throughout the Cambodian Mekong. The results, which were published in the Cambodian Journal of Natural History (Eschenroeder et al. 2024), highlight the potential value that eDNA may provide for fish monitoring in the region, as well as knowledge gaps that still need to be addressed.

Collecting an eDNA sample from the Sekong River.
In 2022, FISHBIO scientists collected 56 eDNA samples from sites in the mainstem Mekong River, its major tributaries (the Sekong, Sesan, and Srepok rivers), and the Tonle Sap (Southeast Asia’s largest lake). Samples were collected by filtering water using eDNA sampling kits from partner laboratory Jonah Ventures, and were then shipped to the US for DNA extraction and sequencing. Though some eDNA approaches are focused on detecting a single species, the method used in this study employed a process called “metabarcoding,” which detects segments of DNA from all fish species present in the collected samples. These DNA sequences are then compared to a reference library of known DNA sequences from a large list of fish species known to occur in the region, allowing for identification of the species that the DNA in the sample originated from.

YEA staff member Sothearoth Chea pushes water through an eDNA collection filter.
The collected samples contained DNA belonging to 63 different types of fish, representing individuals from eight different orders, 23 different families, 49 different genera, and at least 55 species (sometimes sequences could only be identified to the genus level). Among these were several threatened and endangered species, including critically endangered giant barb (Catlocarpio siamensis), critically endangered Mekong giant catfish (Pangasianodon gigas), endangered striped catfish (P. hypophthalmus), threatened small scaled mud carp (Cirrhinus microlepis), and threatened Bandan sharp-mouth barb (Scaphognathops bandanensis). Further, sequences belonging to cryptic, hard to sample fish like loaches (Schistura species) were also detected.

The muddy waters of the Tonle Sap, Southeast Asia’s largest lake. High turbidity in some locations resulted in the filters clogging more quickly, leading to smaller sample volumes.
Although results were promising, the diversity represented in the eDNA samples was only a fraction of what might be expected to occur where the samples were collected. This suggests it may be necessary to filter larger volumes of water to detect the full range of species present in a sample site. This was evident as analysis showed an increase in species detections with increasing volume of water filtered through the collection kits. However, it can be challenging to filter large volumes in certain habitats. The Tonle Sap, for example, has very muddy water, which rapidly clogs the collection filters. Another reason for reduced diversity detected in eDNA samples are gaps in the available DNA sequences for species in the basin. Because of the large number of species present in the Mekong, many species native to the region have not had their DNA sequenced, and therefore there is no way to identify their DNA in eDNA samples. However, efforts to obtain sequences for all Mekong fish species are underway.
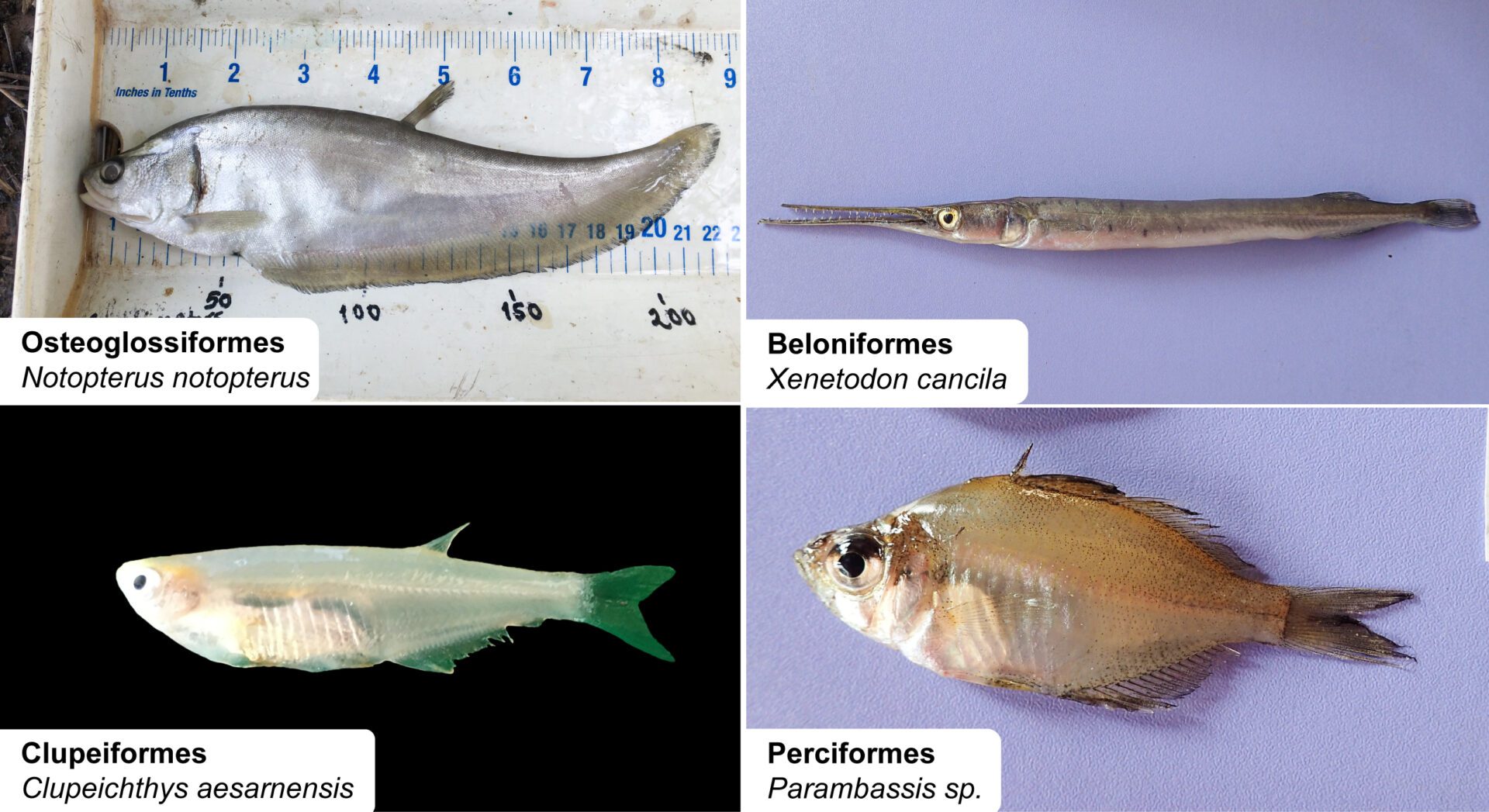

Species belonging to the eight different families represented in the eDNA samples.
This project provided valuable insight into eDNA’s ability to quantify fish diversity and detect threatened and endangered species in the Mekong Basin. The next step in testing the effectiveness of eDNA will require direct comparisons between the number of species detected from eDNA sampling and the number of species detected via traditional, capture-based fish sampling. FISHBIO and project partners at the Royal University of Agriculture are working on a comparative eDNA and traditional sampling project to address this very need, with the analysis of over 200 eDNA samples collected from multiple locations throughout the Cambodian Mekong already underway.

The Sesan River, one of the three major Mekong tributaries sampled for this project.
Standardized eDNA sampling may have numerous applications in the Mekong, including detecting species diversity in areas not represented in current Mekong monitoring programs, and performing biodiversity inventories for environmental impact assessments. Given the immense economic, cultural, and ecological value of fish diversity in the Mekong, and the urgent need for reliable monitoring in the face of biodiversity loss, eDNA may someday soon become an essential tool for monitoring biodiversity and informing conservation approaches.
Header Image: An eDNA filter is capped for storage and transport.
This post was featured in our weekly e-newsletter, the Fish Report. You can subscribe to the Fish Report here.
