


The use of environmental DNA (eDNA) in the Mekong River basin is of increasing interest because of its potential utility for fisheries monitoring and informing natural resource management and conservation of imperiled species. FISHBIO partnered with Young Eco and the Inland Fisheries Research and Development Institute of the Cambodian Fisheries Administration to conduct a pilot study by collecting eDNA samples across the mainstem Mekong, the Sesan, Sekong, and Srepok (3S) tributaries, as well as the Tonle Sap (Southeast Asia’s largest lake) in the spring of 2022. The goal of this pilot study was to assess how much fish biodiversity could be detected using eDNA. Promising results from this effort include the identification of DNA from 63 fish taxa, including critically endangered Mekong giant catfish (Pangasianodon gigas) and Mekong giant barb (Catlocarpio siamensis), endangered striped catfish (Pangasianodon hypophthalmus), and vulnerable Bandan sharp-mouthed barb (Scaphognathops bandanensis) and small scaled mud carp (Cirrhinus microlepis). The results of this pilot effort were published in the December 2023 issue of the Cambodian Journal of Natural History (Eschenroeder et al. 2024).

Fish are harvested on a Dai net platform in the Tonle Sap River.
However, much work remains to be done in terms of evaluating how eDNA can be utilized to assess and monitor Mekong fish species diversity. FISHBIO researchers have been hard at work on a new collaborative effort with the Royal University of Agriculture (RUA) in Phnom Penh, Cambodia – this effort, which is funded by USAID’s Wonders of the Mekong project, is focused on pairing eDNA sample collection with three different fish catch monitoring programs. These include dai net fishery monitoring on the Tonle Sap River, arrow trap monitoring on the Tonle Sap Lake, and experimental gill net sampling in the northern Cambodian Mekong and 3S Basin. Pairing catch from these traditional gears with repeated eDNA sampling will allow for a comparison of the species detected between methods. This is a necessary first step in evaluating eDNA for biodiversity monitoring and is needed in the Mekong system.

Arrow traps in the Tonle Sap viewed from overhead.
Traditional physical fish sampling gears may be selective or limited in their ability to capture the full fish community, and existing monitoring programs occur in only a few specific locations. Therefore, it is critical to evaluate alternative, scalable, and repeatable methods for fisheries monitoring, particularly given the potential fishery impacts of climate change, human alteration of the landscape (e.g., dams, land development), and the cultural, ecological, and economic importance of fisheries in the region. This study’s primary focus is to investigate the feasibility of using eDNA as a monitoring tool, and specifically to evaluate how the biodiversity data generated by eDNA metabarcoding compares to that provided by traditional, capture-based fish monitoring programs. Digging into this question will be essential for understanding effective eDNA applications in the Mekong.
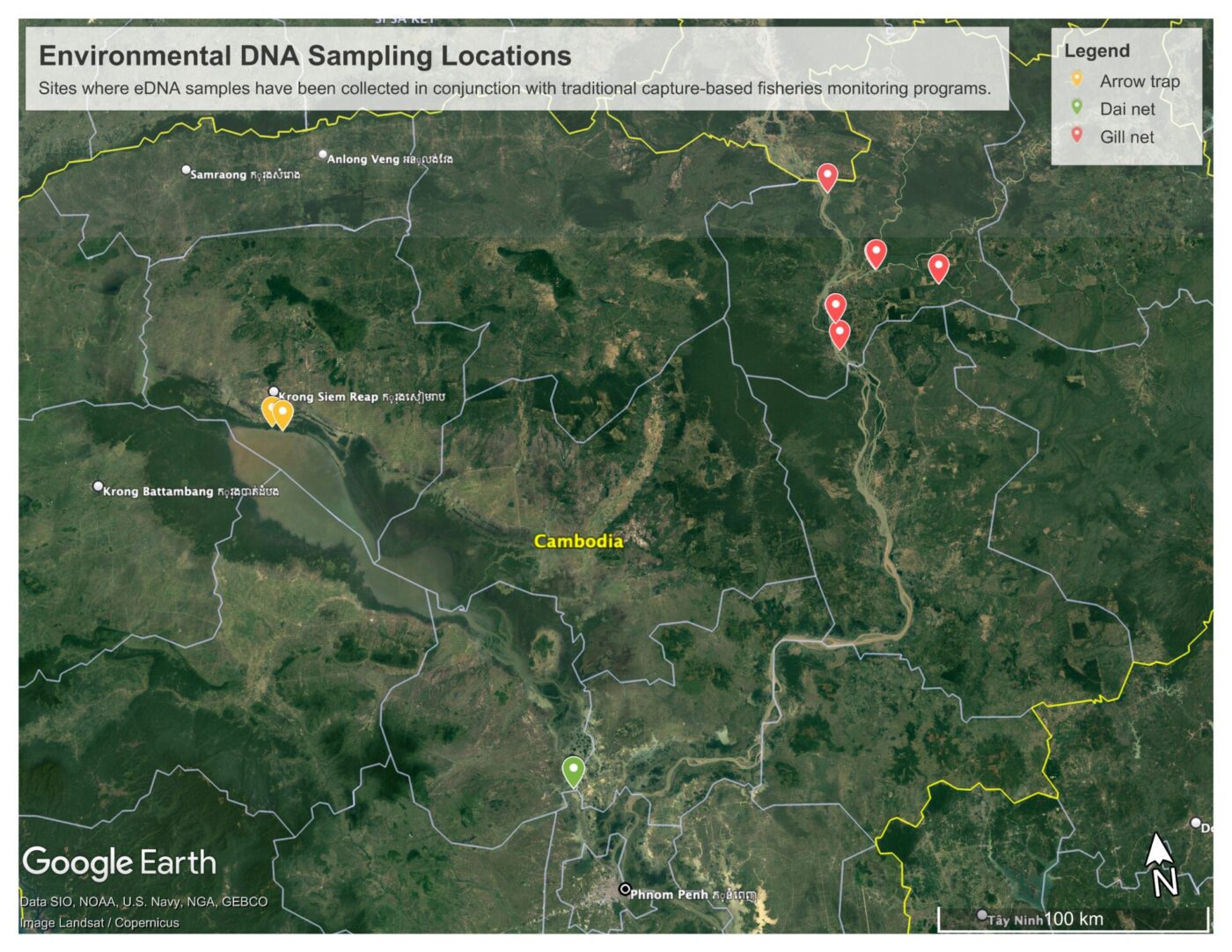
The locations where paired eDNA and traditional sampling are occurring for this project.
This study will provide the first direct comparison between eDNA and capture-based fisheries monitoring approaches in the Lower Mekong Basin, and will generate critical insight into the types of data that eDNA is able to provide, and what types of questions related to fish diversity it may be used to answer. With this knowledge, FISHBIO and project partners hope to contribute to achieving effective use of eDNA in aquatic biodiversity monitoring in Cambodia and beyond.

Gill net sampling in the Mekong in northern Cambodia.
Sample collection began in November of 2022 and continued through October of 2023, covering a full year of fisheries monitoring. Using single-use eDNA filter kits provided by Jonah Ventures (Boulder, Colorado, USA), samples were collected by filtering water at the same locations where the capture-based monitoring was occurring. These samples were collected monthly during the operation of the dai nets from November–February, every month throughout the year alongside arrow trap sampling, and every other month throughout the year alongside experimental gill net sampling. In total, more than 200 samples were collected over the course of the study period, and sample sequencing has begun to reveal promising results. Over the coming months, RUA and FISHBIO scientists will work to analyze the sequence data and compare it with the diversity of species captured by the nets and traps.
Header Image: PhD student Soksan Chhorn collects an eDNA sample on the Sesan River.