Monday May 18, 2015
In a recent Fish Report, we outlined some of the concepts behind aquatic environmental DNA (eDNA) research, and some of the many questions this technique can help answer. While potential uses of eDNA are numerous, unfortunately the procedure isn’t quite as simple as collecting some water and running it through an analysis machine. A wide range of environmental factors, as well as the collection and analysis process, can all influence the results of an eDNA study. Understanding these limitations is important for interpreting the results of this technology, and what it can say about the presence or absence of particular species.
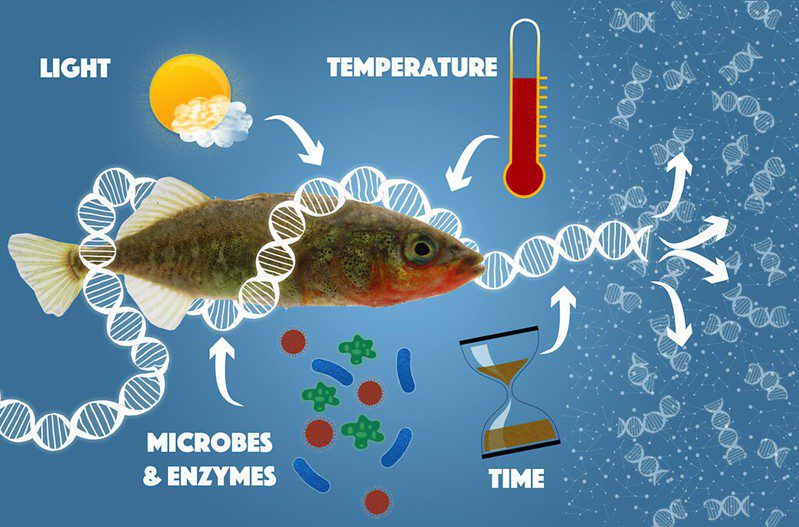
Time is particularly of the essence when it comes to detecting an animal, such as fish, in the environment through eDNA. Once a cell is shed, the animal’s DNA rapidly degrades: a host of microbes and enzymes, as well as environmental factors like light, heat, oxygen, and even water, all work to break up DNA strands into shorter and shorter pieces (Lindahl 1993). After some time, these degraded fragments become too short to provide a unique “genetic fingerprint” to identify a particular species. This can call into question whether that animal is actually present in that environment, or can cause the animal to remain undetected. In the aquatic environment, sturgeon DNA has been shown to persist for up to two weeks in small freshwater ponds, although the probability of detection diminishes over time (Dejean et al. 2011). However, detectability can vary from place to place, and from species to species. For example, salamander eDNA was no longer detectable in a stream within one hour of removing the animals (Pilliod et al. 2013), or after 1-2 weeks in controlled laboratory experiments in an aquarium (Thomsen et al. 2012).
In addition to its short-lived nature, how the animal’s DNA is distributed in the environment also strongly affects the interpretation of eDNA data. In small ponds, evidence suggests that eDNA is evenly distributed in the water, so sampling water anywhere could detect the animal (Thomsen et al. 2012). However, in larger lakes or bays the concentration of eDNA (and therefore its detectability) will decrease with further distances from the animal (Takahara et al. 2013). In rivers, eDNA gets transported downstream, so sampling upstream of an animal will not reveal it. In addition, the likelihood of detection severely diminishes with distance downstream from the animal. For example, the experiment with stream-dwelling salamanders mentioned above found their DNA could be detected 5 meters downstream of the (caged) amphibians, but not 50 meters downstream (Pilliod et al. 2013). In contrast, daphnia eDNA has been reported more than 12 kilometers downstream from the source population (Deiner and Altermatt 2014), which highlights the need to evaluate eDNA sampling capabilities and limitations on a case-by-case (and species-by-species) basis.
Another challenge to interpreting eDNA data is not being able to tell whether the detected animal was alive or dead, and how the eDNA arrived at its location. Pieces of eDNA can be carried from one place to another and create a “false positive,” or an impression that the species is alive and present when it actually is not. Fragments of eDNA can be transported through bilge water discharge on ships, in household wastewater, or even in the excrement of predatory species that consumed their prey elsewhere. False positive detections from these transport events could in turn potentially trigger managers to mount costly and unnecessary eradication efforts to curb the spread of the phantom invasive species (a topic of ongoing controversy regarding spread of Asian carp to the Great Lakes from Mississippi River basin; Wittmann et al. 2014).
Ultimately, the detectability of eDNA depends on the size and number of a particular organism, and the rates at which DNA is shed by the organisms and degraded in the environment. While eDNA is increasingly being used to supplement traditional sampling and observational studies, significant challenges still remain to using this technology reliably, and to learn more than simply whether an animal is present or absent. It appears that eDNA methods can provide useful tools to help scientists improve our understanding of where species live and travel, but it’s important to carefully consider the limitations and context when applying this exciting technology (Bohmann et al. 2014). We’ll talk about these applications in a future Fish Report – stay tuned!This post featured in our weekly e-newsletter, the Fish Report. You can subscribe to the Fish Report here.
