Monday April 22, 2024
Humans have developed an untold number of ways to catch fish, but no matter how many traps, electrofishers, snorkel surveys, and even good ol’ fashioned fishing poles a fisheries biologist may employ, these sampling methods never guarantee the capture of every individual. This can make it difficult to obtain a complete assessment of a fish community. But while fish may evade fykes, hooks, and nets, they still leave telltale traces in the environment in the form of DNA. Collecting environmental DNA (eDNA) is a promising modern approach to fisheries sampling, and has the potential to revolutionize the way fisheries biologists measure aquatic biodiversity. Metabarcoding and qPCR have emerged as two particularly effective techniques for eDNA sample analysis that are capable of detecting species present in a water body from just a sample of water, making it harder for rare and wily fish to go undetected.

Collecting an eDNA sample from a stream with a single-use collection kit.
Out in the field, obtaining an eDNA sample can be as simple as filtering water with a single-use syringe kit. The syringe is used to draw up a water sample, and then to push that water through a filter that traps genetic material suspended in the water. Other methods of collection are similar in concept and process, but use automated or manual pumps to filter larger volumes of water. These vary from commercially available devices to DIY. versions that can be constructed from materials available at a local hardware store. Back in the lab, geneticists extract DNA fragments from the filter, and can detect and identify the DNA present in the sample using a variety of methods. The two most broadly applied analysis techniques in fisheries research are known as quantitative polymerase chain reaction (qPCR) and metabarcoding. Each procedure provides different information that can be used to answer different research questions.

Water is pushed through a very fine filter to capture particles that may contain DNA.
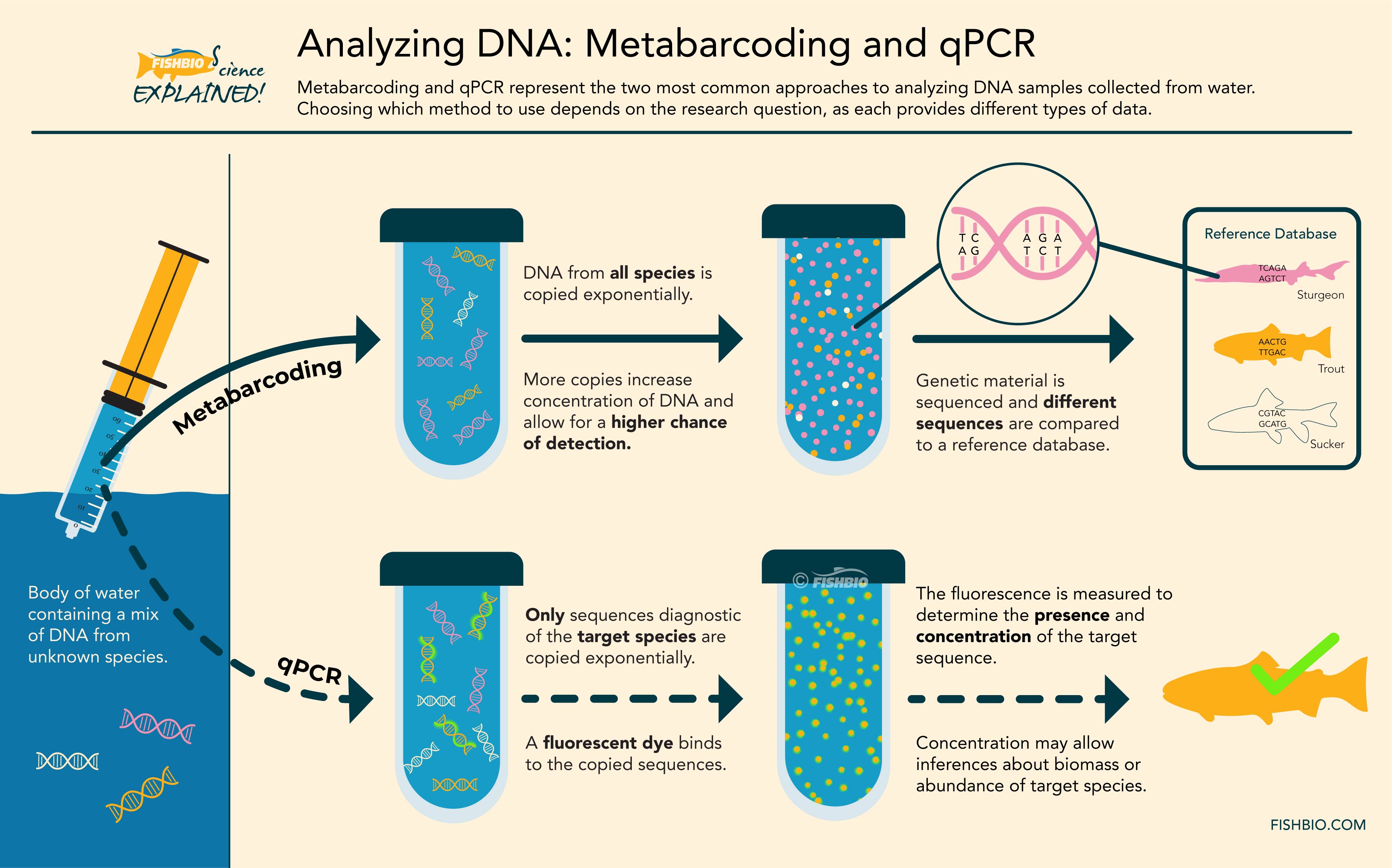
Both qPCR and metabarcoding require the extracted DNA to undergo an enzymatic reaction referred to as polymerase chain reaction (or PCR), which amplifies the DNA present in the sample, resulting in thousands of copies. A biologist interested in a single target species (as may be the case when monitoring at-risk or invasive species) may choose to apply the qPCR technique, during which only DNA from the target organism is copied. By incorporating fluorescent dye into the PCR reaction – and therefore in each copy of the target DNA – geneticists can measure the amount of DNA belonging to the species of interest that is present in the sample. In contrast, metabarcoding uses PCR to amplify DNA from many species present in the sample, which are then sequenced and compared to a reference database containing genetic sequences of already-identified species. This technique can facilitate the identification of hundreds of species in a single sample. As such, metabarcoding is generally used when scientists aim to evaluate the entire fish community in a location, and is well suited for estimating the diversity of fish species in a given area. Metabarcoding can also allow for analysis of stomach samples to study fish diets, or for detection of other living things in the aquatic environment, from reptiles to amphibians to plankton.

Preservative solution is added to the filter to stabilize DNA until it is extracted in the lab.
Every biological compound breaks down over time, and DNA is no exception. Between degradation, movement with current, or dilution in large water bodies, detecting DNA can be a bit like finding a needle in a haystack – especially when searching for rare species. Researchers have discovered that, generally, DNA particles remain intact enough to be analyzed for about a week before the window for successful detection begins to close , requiring relatively recent presence and close proximity of sampling to the target species for it to be detected. Furthermore, species detection through the metabarcoding approach is only as complete as the reference databases that geneticists have to identify samples.
While in the field collecting eDNA samples, it is important to record data related to sample collection, including date, time, location, and volume filtered.
With the ability to replace hours of field effort and loads of sampling gear with a few minutes of time and a sample of water, it is easy to understand why eDNA sampling is gaining popularity. A fish community assessment method that negates the need to capture and handle fish is valuable to fisheries researchers studying rare and endangered species. However, this technology is still relatively new, and despite reported success and accuracy amongst scientists, it is not without nuance. It is critical to remember that while eDNA can provide valuable data on diversity, species distributions, and maybe even abundance in certain cases, traditional capture-based fish sampling will continue to be essential to collect important data on individual fish, including sex, age, diet, and size. As such, eDNA is best applied as a complementary approach to traditional monitoring and research. But as molecular technology continues to improve and geneticists gradually add more fish species to existing genetic reference databases, eDNA technology will only grow in utility for modernized fisheries monitoring.
Header Image: A diagram of the processes involved in metabarcoding and qPCR.
This post was featured in our weekly e-newsletter, the Fish Report. You can subscribe to the Fish Report here.
